Note
Go to the end to download the full example code.
getting started#
demonstration of basic pymiediff functionality:
Extinction, scattering and absorption spectra
Angular scattering patterns
author: P. Wiecha, 03/2025
imports#
import numpy as np
import torch
import matplotlib.pyplot as plt
from matplotlib.patches import Circle
import pymiediff as pmd
setup#
we setup the particle dimension and materials as well as the environemnt. This is then wrapped up in an instance of Particle.
# - config
wl0 = torch.linspace(500, 1000, 100)
k0 = 2 * torch.pi / wl0
r_core = 70.0
r_shell = 100.0
mat_core = pmd.materials.MatDatabase("Si")
mat_shell = pmd.materials.MatDatabase("Ge")
n_env = 1.0
# - setup the particle
p = pmd.Particle(
r_core=r_core,
r_shell=r_shell,
mat_core=mat_core,
mat_shell=mat_shell,
mat_env=n_env,
)
print(p)
core-shell particle
- core radius = 70.0nm
- shell radius = 100.0nm
- core material : Si
- shell material : Ge
- environment : eps=1.00
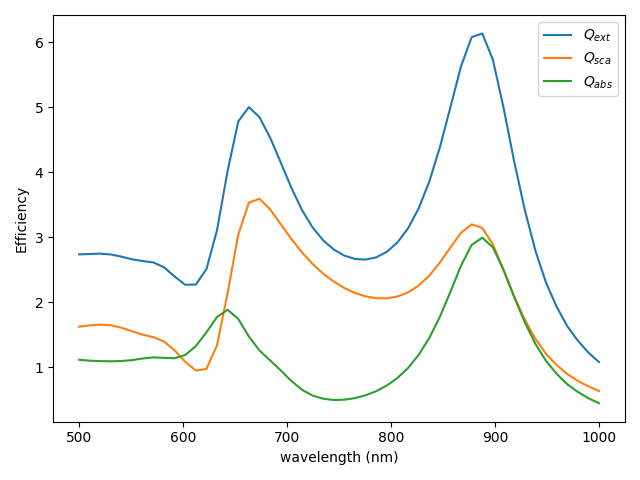
efficiency spectra#
Calculate extinction, scattering and absorption spectra
cs = p.get_cross_sections(k0)
# - plot
plt.figure()
plt.plot(cs["wavelength"], cs["q_ext"], label="$Q_{ext}$")
plt.plot(cs["wavelength"], cs["q_sca"], label="$Q_{sca}$")
plt.plot(cs["wavelength"], cs["q_abs"], label="$Q_{abs}$")
plt.xlabel("wavelength (nm)")
plt.ylabel("Efficiency")
plt.legend()
plt.tight_layout()
# plt.savefig("ex_01a.svg", dpi=300)
plt.show()
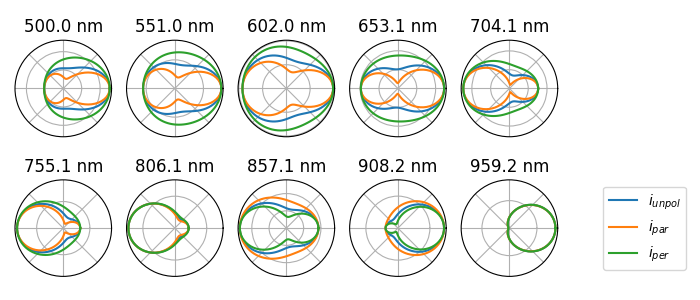
angular scattering#
Calculate angular radiation patterns of the scattering
theta = torch.linspace(0.0, 2 * torch.pi, 100)
angular = p.get_angular_scattering(k0, theta)
# - plot every 10th wavelength
plt.figure(figsize=(7, 3))
for i, i_k0 in enumerate(range(len(k0))[::10]):
ax = plt.subplot(2, 5, i + 1, polar=True)
plt.title(f"{wl0[i_k0]:.1f} nm")
ax.plot(angular["theta"], angular["i_unpol"][i_k0], label="$i_{unpol}$")
ax.plot(angular["theta"], angular["i_par"][i_k0], label="$i_{par}$")
ax.plot(angular["theta"], angular["i_per"][i_k0], label="$i_{per}$")
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.legend(loc="center left", bbox_to_anchor=(1.4, 0.5))
plt.tight_layout()
# plt.savefig("ex_01b.svg", dpi=300)
plt.show()
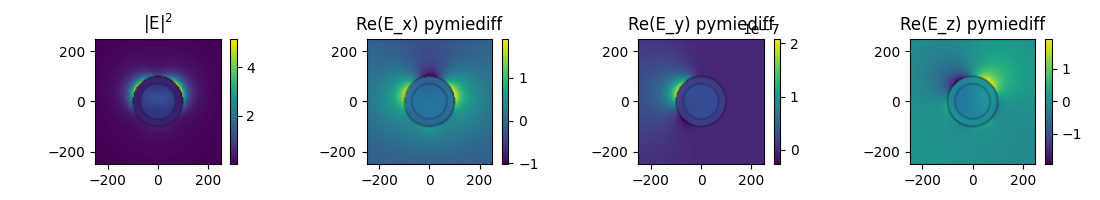
nearfields#
Calculate nearfields in and around the partilce
# create grid of evaluation positions
d_area_plot = 250.0
x, z = torch.meshgrid(
torch.linspace(-d_area_plot, d_area_plot, 100),
torch.linspace(-d_area_plot, d_area_plot, 100),
)
y = torch.zeros_like(x)
grid = torch.stack([x, y, z], dim=-1)
orig_shape_grid = grid.shape
# evaluate the fields
i_wl = 15
fields_results = p.get_nearfields(k0=k0[i_wl], r_probe=grid.view(-1, 3))
# reshape fields to 2D grid
E_s = fields_results["E_s"]
E_s = E_s.view(orig_shape_grid)
intensity_sca_mie = torch.sum(E_s.abs() ** 2, dim=-1)
# --- plot
def plot_circles(radii, im=None):
for r in radii:
rect = Circle(
xy=[0, 0],
radius=r,
linewidth=1.5,
edgecolor="k",
facecolor="C0",
alpha=0.25,
)
plt.gca().add_patch(rect)
if im is not None:
cb = plt.colorbar(im, label="")
fig = plt.figure(figsize=(11, 2))
# - intensity
plt.subplot(141, aspect="equal", title="|E|$^2$")
im = plt.imshow(
intensity_sca_mie.T,
extent=(2 * (-d_area_plot, d_area_plot)),
origin="lower",
)
plot_circles([r_core, r_shell], im)
# - field comp. (real part)
Ecomp_str = ["x", "y", "z"]
for i in range(3):
plt.subplot(1, 4, i + 2, aspect="equal", title=f"Re(E_{Ecomp_str[i]}) pymiediff")
im = plt.imshow(
E_s[..., i].T.imag,
extent=(2 * (-d_area_plot, d_area_plot)),
origin="lower",
)
plot_circles([r_core, r_shell], im)
plt.tight_layout()
plt.show()
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/torch/functional.py:505: UserWarning: torch.meshgrid: in an upcoming release, it will be required to pass the indexing argument. (Triggered internally at /pytorch/aten/src/ATen/native/TensorShape.cpp:4317.)
return _VF.meshgrid(tensors, **kwargs) # type: ignore[attr-defined]
Total running time of the script: (0 minutes 2.230 seconds)
Estimated memory usage: 668 MB
