Note
Go to the end to download the full example code.
benchmark#
Direct Mie evluation benchmark against other tools
author: P. Wiecha, 09/2025
imports#
import time
import numpy as np
import matplotlib.pyplot as plt
import torch
import pymiediff as pmd
# other Mie tools
import pymiecs as pmc
import treams
from TypedUnit import ureg
from PyMieSim.experiment.scatterer import CoreShell
from PyMieSim.experiment.source import Gaussian
from PyMieSim.experiment import Setup
setup test case#
we setup a simple test case
# - config
N_wl = 256
wl0 = torch.linspace(500, 1500, N_wl)
k0 = 2 * torch.pi / wl0
r_core = 200.0
r_shell = 420.0
n_core = 0.5 + 5.1j
n_shell = 4.5 # caution: metal-like shells can become unstable
mat_core = pmd.materials.MatConstant(n_core**2)
mat_shell = pmd.materials.MatConstant(n_shell**2)
n_env = 1.0
Mie from treams#
we benchmark against the t-matrix toolkit treams: tfp-photonics/treams write a simple wrapper for the treams Mie code.
def mie_ab_sphere_treams(k0: np.ndarray, radii: list, materials: list, n_env, n_max):
"""Mie scattering via package `treams`, for vacuum environment"""
assert len(radii) == len(materials)
# embedding medium: vacuum
eps_env = n_env**2
# main Mie extraction and setup
a_n = np.zeros((len(k0), n_max), dtype=np.complex128)
b_n = np.zeros_like(a_n)
for i_wl, _k0 in enumerate(k0):
# core and shell materials
mat_treams = []
for eps_mat in materials:
mat_treams.append(treams.Material(eps_mat))
# treams convention: environment material last
mat_treams.append(treams.Material(eps_env))
for n in range(1, n_max + 1):
miecoef = treams.coeffs.mie(n, _k0 * np.array(radii), *zip(*mat_treams))
a_n[i_wl, n - 1] = -miecoef[0, 0] - miecoef[0, 1]
b_n[i_wl, n - 1] = -miecoef[0, 0] + miecoef[0, 1]
# scattering
Qs_mie = np.zeros(len(k0))
for n in range(n_max):
Qs_mie += (2 * (n + 1) + 1) * (np.abs(a_n[:, n]) ** 2 + np.abs(b_n[:, n]) ** 2)
Qs_mie *= 2 / (k0 * n_env * max(radii)).real ** 2
return dict(
a_n=a_n,
b_n=b_n,
q_sca=Qs_mie,
wavelength=2 * np.pi / k0,
)
t0 = time.time()
cs_treams = mie_ab_sphere_treams(
k0=k0.numpy(),
radii=[r_core, r_shell],
materials=[n_core**2, n_shell**2],
n_env=n_env,
n_max=15,
)
t_treams = time.time() - t0
PyMieSim#
https://martinpdes.github.io/PyMieSim/
# defining source and particle
source = Gaussian(
wavelength=wl0.cpu().numpy() * ureg.nanometer,
polarization=0 * ureg.degree,
optical_power=1e-3 * ureg.watt,
NA=0.5 * ureg.AU,
)
scatterer = CoreShell(
core_diameter=2 * r_core * ureg.nanometer, # Core diameters from 100 nm to 600 nm
shell_thickness=(r_shell - r_core) * 2 * ureg.nanometer, # Shell width of 800 nm
core_property=n_core * ureg.RIU, # Core material
shell_property=n_shell * ureg.RIU, # Shell material
medium_property=n_env * ureg.RIU, # Surrounding medium's refractive index
source=source,
)
# "experiment" setup
experiment = Setup(scatterer=scatterer, source=source)
# Measuring the properties
t0 = time.time()
dataframe = experiment.get("Qsca", scale_unit=True)
q_sca_pms = np.array([float(f[0]) for f in dataframe["Qsca"].to_numpy()])
t_pms = time.time() - t0
PyMieCS#
# - calculate efficiencies
t0 = time.time()
cs_pmc = pmc.Q(
k0,
r_core=r_core,
n_core=n_core,
r_shell=r_shell,
n_shell=n_shell,
n_env=n_env,
n_max=15,
)
t_pymiecs = time.time() - t0
PyMieDiff#
Differentiable Mie
# - setup the particle
p = pmd.Particle(
r_core=r_core,
r_shell=r_shell,
mat_core=mat_core,
mat_shell=mat_shell,
mat_env=n_env,
)
# - scipy wrapper for Mie coefficients
t0 = time.time()
cs_pmd = p.get_cross_sections(k0, backend="scipy")
t_pymiediff_scipy = time.time() - t0
# - native torch Mie coefficients
t0 = time.time()
cs_pmd_torch = p.get_cross_sections(k0, backend="torch")
t_pymiediff_torch = time.time() - t0
# - native torch + batched evaluation
N_batch = 128
r_c_many = torch.linspace(r_core, r_core + 50, N_batch)
r_s_many = torch.linspace(r_shell, r_shell + 50, N_batch)
eps_c_many = n_core**2 + torch.linspace(0, 1, N_batch).unsqueeze(0).broadcast_to(
N_wl, N_batch
)
eps_s_many = n_shell**2 + torch.linspace(0, 1, N_batch).unsqueeze(0).broadcast_to(
N_wl, N_batch
)
t0 = time.time()
res_mie = pmd.coreshell.cross_sections(
k0,
r_c=r_c_many,
eps_c=eps_c_many,
r_s=r_s_many,
eps_s=eps_s_many,
eps_env=n_env**2,
backend="torch",
)
t_pymiediff_batch = (time.time() - t0) / (N_batch)
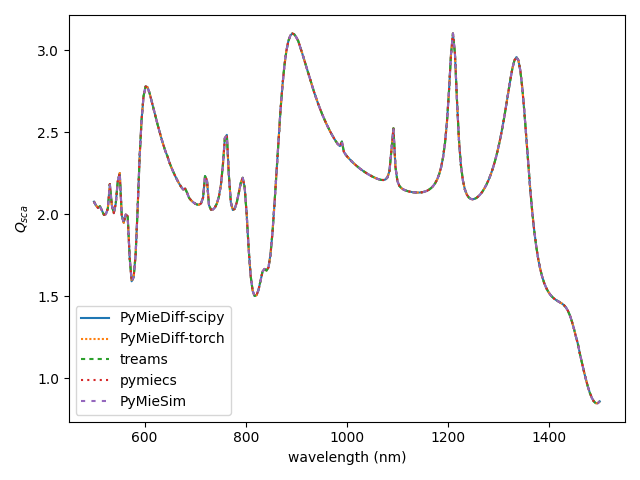
plot spectra of different tools#
plt.figure()
plt.plot(cs_pmd["wavelength"], cs_pmd["q_sca"], label="PyMieDiff-scipy")
plt.plot(
cs_pmd["wavelength"], cs_pmd_torch["q_sca"], label="PyMieDiff-torch", dashes=[1, 1]
)
plt.plot(cs_treams["wavelength"], cs_treams["q_sca"], label="treams", dashes=[2, 2])
plt.plot(cs_pmd["wavelength"], cs_pmc["qsca"], label="pymiecs", dashes=[1, 2])
plt.plot(cs_pmd["wavelength"], q_sca_pms, label="PyMieSim", dashes=[2, 3])
plt.xlabel("wavelength (nm)")
plt.ylabel("$Q_{sca}$")
plt.legend()
plt.tight_layout()
plt.show()
timing results#
toolkits = [
"treams",
"PyMieSim",
"PyMieCS",
"PyMieDiff\n(scipy)",
"PyMieDiff\n(torch)",
"PyMieDiff\n(batched)",
]
timing = [
t_treams * 1e6 / N_wl,
t_pms * 1e6 / N_wl,
t_pymiecs * 1e6 / N_wl,
t_pymiediff_scipy * 1e6 / N_wl,
t_pymiediff_torch * 1e6 / N_wl,
t_pymiediff_batch * 1e6 / N_wl,
]
bar_colors = ["C0", "C1", "C2", "C4", "C5", "C6"]
fig, ax = plt.subplots(figsize=(4.5, 3.5))
ax.bar(toolkits, timing, color=bar_colors)
plt.xticks(rotation=45)
for i, t in enumerate(timing):
plt.text(
i,
plt.ylim()[1] * 0.8,
r"{:.1f}$\,$µs".format(t),
rotation=90,
va="top",
ha="center",
)
ax.set_ylabel(r"time per wavelength (µs)")
ax.set_title("")
plt.tight_layout()
plt.show()
# - print timing results
print("calculated {} wavelengths.".format(N_wl))
print(50 * "-")
print(
"time PyMieDiff (torch): {:.4f} ms / wl".format(
(t_pymiediff_torch) * 1e3 / N_wl
)
)
print(
"time PyMieDiff (torch, batched): {:.4f} ms / wl".format(
(t_pymiediff_batch) * 1e3 / N_wl
)
)
print(
"time PyMieDiff (scipy): {:.4f} ms / wl".format(
(t_pymiediff_scipy) * 1e3 / N_wl
)
)
print(50 * "-")
print(
"time pymiecs: {:.4f} ms / wl".format((t_pymiecs) * 1e3 / N_wl)
)
print("time pymiesim: {:.4f} ms / wl".format((t_pms) * 1e3 / N_wl))
print("time treams: {:.4f} ms / wl".format((t_treams) * 1e3 / N_wl))
print(50 * "-")
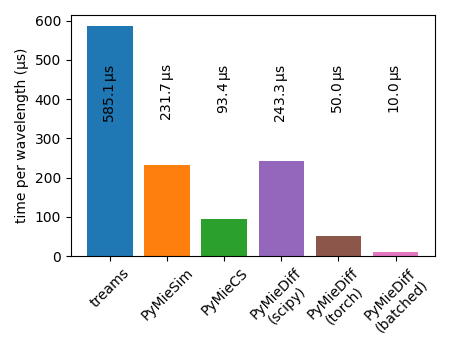
calculated 256 wavelengths.
--------------------------------------------------
time PyMieDiff (torch): 0.0335 ms / wl
time PyMieDiff (torch, batched): 0.0104 ms / wl
time PyMieDiff (scipy): 0.1021 ms / wl
--------------------------------------------------
time pymiecs: 0.0949 ms / wl
time pymiesim: 0.2455 ms / wl
time treams: 0.5927 ms / wl
--------------------------------------------------
Total running time of the script: (0 minutes 3.070 seconds)
Estimated memory usage: 1230 MB
